In response to several blog posts earlier on this semester, describing our course’s broad application to biological networks (such as this one, this one, and this one) I highlight that Piers J Ingram, Michael PH Stump and Jaroslav Stark published research in 2006 demonstrating that network effects in biology are not only often poorly understood, but in their example are also surprising in the manner which they fail to elucidate function in their larger systems. The article, available here , describes the simulated dynamics of a bi-fan motif found in the transcription regulation network of S. cerevisiae (baking yeast). Nodes represent genes, and the directed edges between them represent how the protein products of the genes regulate the transcription- the process in which the information in DNA is copied to RNA- of other genes. Figure B, as taken from the article, below, is known as the bi-fan motif, where a motif is defined as a particular node-edge structure that is found with particularly high frequency in any network, relative to a “null hypothesis” with that network constructed with random edges.
, describes the simulated dynamics of a bi-fan motif found in the transcription regulation network of S. cerevisiae (baking yeast). Nodes represent genes, and the directed edges between them represent how the protein products of the genes regulate the transcription- the process in which the information in DNA is copied to RNA- of other genes. Figure B, as taken from the article, below, is known as the bi-fan motif, where a motif is defined as a particular node-edge structure that is found with particularly high frequency in any network, relative to a “null hypothesis” with that network constructed with random edges.
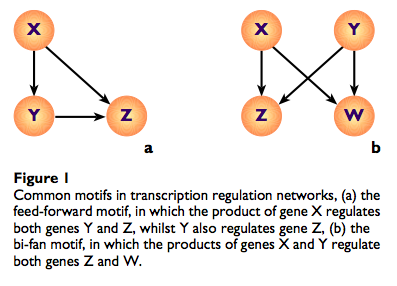
By using differential equations to describe the concentrations, over time, of the relevant molecules in this network found in S. cerevisiae, these researchers were able to simulate how the molecules in this network would be produced. What they found was that depending on the particular genes they chose, there was, in fact, no set of consistent regulatory behavior that the bi-fan motif exhibited.
This is a curious result, for several reasons. First off, although essentially what’s happening here is a set of interactions between genes and their protein products which regulate other genes- a situation that ostensibly lends itself naturally to analysis by networks- this scenario seems to not be easily modeled by networks, or at least by network motifs. Networks are not enough. Rather, a priori biological knowledge about the situation is required to generate an accurate model. Furthermore, and perhaps surprisingly, this implies that the motif structure of these small networks do not define their function. In contrast, as oft repeated as a theme in introductory biology courses, and as is found true in much research, biological structure determines biological function.
Despite what shortcomings this work by Ingram et al. may have in terms of network modeling, it certainly provides a valuable, abstract suggestion to biological reductionists interested in interactions between components of biological systems: perhaps structure does not indeed always determine function.
—–
The article discussed in this post can be found as: Barabasi, Albert-Laszlo and Zoltan N. Oltvai. “Network motifs: structure does not determine function.” BMC Genomics 7:108 (2006): 28 February 2008 <http://www.biomedcentral.com/1471-2164/7/108>


* You can follow any responses to this entry through the RSS 2.0 feed.